Dr. Xiaoyu Xue
Associate Professor

Office: CENT 404
Laboratory: CENT 406A
Phone: (512) 245-6607
Fax: (512) 245-2374
email: xiaoyu.xue@txstate.edu
Xue Lab Website: https://xuelab.wp.txstate.edu
Educational Background
- Associate Research Scientist, Molecular Biophysics and Biochemistry, Yale University School of Medicine, New Haven, CT (2014-2018)
- Postdoctoral Associate, Molecular Biophysics and Biochemistry, Yale University School of Medicine, New Haven, CT (2008-2014)
- Ph.D., Biology, Tsinghua University, Beijing, China (2008)
- B.S., Biochemistry, Nanjing University, Nanjing, China (2003
Honors & Awards
- Grant Award in the amount of $436,350 from the National Institute of General Medical Sciences of the National Institutes of Health
- Awards of the 10th Challenge Cup (Grade 3), China, 2007
- Hongkong Hengtai Scholarship (Grade 1) of Tsinghua University, China, 2007
- Innovation awards of the 23rd Innovation Contest of Tsinghua University, China, 2005
- Second-class awards of the 23rd Innovation Contest of Tsinghua University, China, 2005
Areas of Interest
- Mechanism of DNA/RNA helicases and nucleases in DNA replication and repair
- DNA replication fork repair
- R-loop processing
- DNA double strand break (DSB) repair and Homologous recombination
- Structural biology
Research in the Xue Group
My previous research focused on understanding conserved mechanisms of DNA replication and repair using methods of biochemistry, genetics, cell biology, and structural biology. In particular, my studies have provided mechanistic information on how the FANCM/Mph1/Fml1 family DNA motor proteins, mutated in the cancer prone disease Fanconi anemia, mediate DNA replication fork repair and the regulation of DNA break repair by homologous recombination. I demonstrated for the first time how the budding yeast FANCM ortholog Mph1 catalyzes replication fork regression and DNA recombination steps, and how its activities are differentially regulated in vitro and in cells by conserved SMC and histone fold complexes (Xue et al., Mol Cell 2014; Xue et al., Genes Dev 2015). I further discovered a new Mph1 partner, Mte1, and defined its roles in regulating Mph1 (Xue et al., Genes & Dev 2016). These studies provide a paradigm for understanding DNA helicase-mediated control of replication fork regression and DNA recombination. These and other research findings are summarized in a review (Xue et al., Genes & Dev 2015).
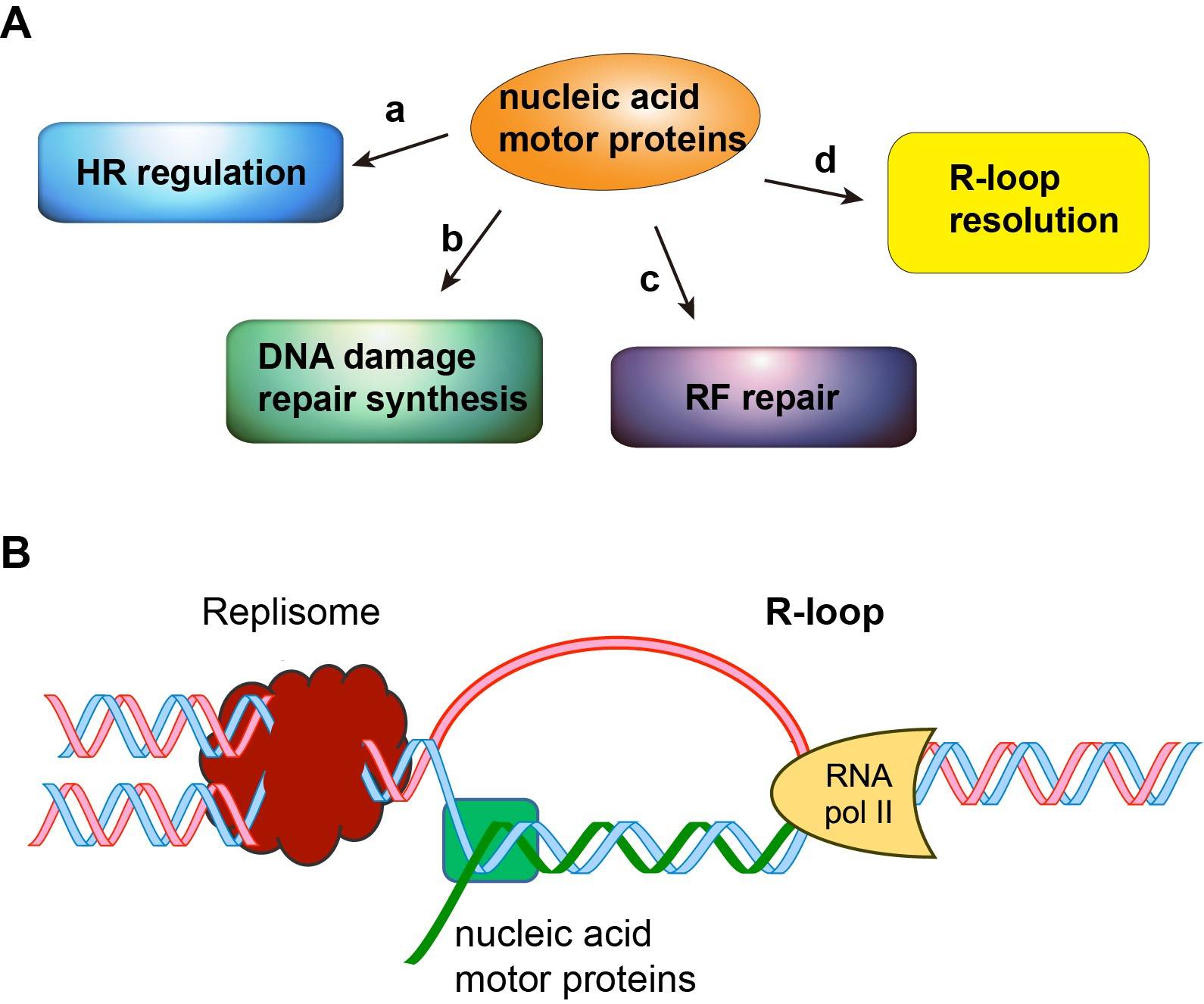
Currently, research in the Xue group is aimed at investigating how DNA/RNA motor proteins enable precise DNA repair to avoid cell transformation and tumorigenesis (Figure). We have obtained important preliminary data on two human helicases and established a strong foundation for mechanistic studies in my laboratory. My research projects include: (1) Roles of nucleic acid motor protein ZGRF1 in chromosome damage repair and disease avoidance (2) Functions of the DNA/RNA motor protein AQR in R-loop resolution. Both lines of studies have great potential for the development of novel strategies to treat human diseases that stem from defects or deregulation in DNA repair, such as Fanconi anemia, neurodegeneration diseases, and familial breast and ovarian cancers. Thus, my studies are both fundamentally important and clinically relevant.
Selected Recent Publications:
Marchena-Cruz, E., Camino, L. P., Bhandari, J. #, Silva, S., Marqueta-Gracia, J. J., Amdeen, S. A. #, Guillén-Mendoza, C., García-Rubio, M. L., Calderón-Montaño, J. M., Xue, X., Luna, R.*, and Aguilera, A*. DDX47, MeCP2 and other functionally heterogeneous factors protect cells from harmful R-loops. Cell Rep. 2023, 42(3): 112148. DOI: 10.1016/j.celrep.2023.112148
Li, S., Mutchler, A. # (co-first author), Zhu, X. #, So, S. #, Epps, J. #, Guan, D., Zhao, X.*, and Xue, X* (co-corresponding author). Multifaceted regulation of the sumoylation of the Sgs1 DNA helicase. J Biol Chem. 2022, 298(7): 102092. DOI: 10.1016/j.jbc.2022.102092
Li, S., Bonner, J. N., Wan, B., So, S. #, Mutchler, A. #, Gonzalez, L. #, Xue, X.*, and Zhao, X.* (co-corresponding author). Esc2 orchestrates substrate-specific sumoylation by acting as a SUMO E2 cofactor in genome maintenance. Genes Dev. 2021, 35(3-4): 261-272. DOI: 10.1101/gad.344739.120
Lisby, M.* and Xue, X*. Protocol for Purification of Human ZGRF1 and Its Regulatory Function on RAD51-Mediated D-Loop Formation. STAR Protoc. 2020, 1(2): 100099. DOI: 10.1016/j.xpro.2020.100099
Brannvoll, A.*, Xue, X.* (co-first author), Kwon, Y., Kompocholi, S., Simonsen, A. K. W., Viswalingam, K. S., Gonzalez, L.#, Hickson, I. D., Oestergaard, V. H., Mankouri, H. W., Sung, P., Lisby, M. The ZGRF1 Helicase Promotes Recombinational Repair of Replication-Blocking DNA Damage in Human Cells. Cell Rep. 2020, 32(1): 107849. DOI: 10.1016/j.celrep.2020.107849
Pérez-Calero, C., Bayona-Feliu, A., Xue, X., Barroso, S. I., Muñoz, S., González-Basallote, V. M., Sung, P., Aguilera, A. UAP56/DDX39B is a major cotranscriptional RNA-DNA helicase that unwinds harmful R loops genome-wide. Genes Dev. 2020, 34(13-14): 898-912. DOI: 10.1101/gad.336024.119
Daley, J. M., Tomimatsu, N., Hooks, G., Wang, W., Miller, A. S., Xue, X., Nguyen, K. A., Kaur, H., Williamson, E., Mukherjee, B., Hromas, R., Burma, S., Sung, P. Specificity of end resection pathways for double-strand break regions containing ribonucleotides and base lesions. Nat Commun. 2020, 11(1): 3088. DOI: 10.1038/s41467-020-16903-4
Xue, C., Daley, J. M., Xue, X., Steinfeld, J., Kwon, Y., Sung, P., Greene, E. C. Single-molecule visualization of human BLM helicase as it acts upon double- and single-stranded DNA substrates. Nucleic Acids Res. 2019, 47(21): 11225-11237. DOI: 10.1093/nar/gkz810
Jiang, H., Xue, X., Panda, S., Kawale, A., Hooy, R. M., Liang, F., Sohn, J., Sung, P., Gekara, N. O. Chromatin-bound cGAS is an inhibitor of DNA repair and hence accelerates genome destabilization and cell death. EMBO J. 2019, 38(21): e102718. DOI: 10.15252/embj.2019102718
Wang, W., Daley, J. M., Kwon, Y., Xue, X., Krasner, D., Miller, A. S., Nguyen, K., Williamson, E., Shim, E., Lee, S., Hromas, R., and Sung, P. A DNA nick at Ku-blocked double-strand break ends serves as an entry site for exonuclease 1 (Exo1) or Sgs1-Dna2 in long-range DNA end resection. J Biol. Chem. 2018, 293(44):17061-17069. DOI: 10.1074/jbc.RA118.004769
Daley, J. M., Jimenez-Sainz, J., Wang, W., Miller, A. S., Xue, X., Nguyen, K. A., Jensen, R. B., and Sung, P. Enhancement of BLM-DNA2-Mediated Long-Range DNA End Resection by CtIP. Cell Report. 2017, 21(2): 324-332. DOI: 10.1016/j.celrep.2017.09.048
Xue, X., Papusha, A., Choi, K., Bonner, J., Niu, H., Kaur, H., Zheng, X. F., Donnianni, R., Lu, L., Lichten, M., Zhao, X., Ira, G., and Sung, P. Differential regulation of the anti-crossover and replication fork regression activities of Mph1 by Mte1. Genes Dev. 2016, 30(6): 687-699. DOI: 10.1101/gad.276139.115
Bonner, J., Choi, K., Xue, X., Torres, N., Szaka, B., Wei, L., Wan, B., Arter, M., Matos, J., Sung, P., Brown, G., Branzei, D., and Zhao, X. Smc5/6 Mediated Sumoylation of the Sgs1-Top3-Rmi1 Complex Promotes Removal of Recombination Intermediates. Cell Report. 2016, 16(2): 368-378. DOI: 10.1016/j.celrep.2016.06.015
Xue, X., Sung, P. and Zhao, X. Functions and regulation of multi-tasking FANCM family of DNA motor proteins (invited review). Genes Dev. 2015, 29(17): 1777-1788. DOI: 10.1101/gad.266593.115
Xue, X.*, Choi, K.* (co-first author), Bonner, J., Szakal, B., Papusha, A., Saro, D., Niu, H., Ira, G., Branzei, D., Sung, P., and Zhao, X. Selective modulation of the functions of a conserved DNA motor by a histone-fold complex. Genes Dev. 2015, 29(10): 1000-1005. DOI: 10.1101/gad.259143.115
Xue, X., Choi, K., Bonner, J., Chiba, T., Kwon, Y., Xu, Y., Sanchez, H., Wyman, C., Niu, H., Zhao, X., and Sung, P. Restriction of Replication Fork Regression Activities by a Conserved SMC Complex. Mol. Cell. 2014, 56(3): 436-445. DOI: 10.1016/j.molcel.2014.09.013
Zhao, Q.*, Xue, X.*, Longerich, S.* (co-first authors), Sung, P., and Xiong, Y. Structural Insights into 5' Flap DNA Unwinding and Incision by the Human FAN1 Dimer. Nat Commun. 2014, 5:5726. DOI: 10.1038/ncomms6726
Lisby M and Xue X, Protocol for Purification of Human ZGRF1 and Its Regulatory Function on RAD51-Mediated D-Loop Formation. STAR Protoc. 2020, 1(2): 100099. DOI: 10.1016/j.xpro.2020.100099
Brannvoll A*, Xue X* (co-first author), Kwon Y, Kompocholi S, Simonsen AKW, Viswalingam KS, Gonzalez L, Hickson ID, Oestergaard VH, Mankouri HW, Sung P, Lisby M. The ZGRF1 Helicase Promotes Recombinational Repair of Replication-Blocking DNA Damage in Human Cells. Cell Rep. 2020, 32(1): 107849. DOI: 10.1016/j.celrep.2020.107849
Pérez-Calero C, Bayona-Feliu A, Xue X, Barroso SI, Muñoz S, González-Basallote VM, Sung P, Aguilera A. UAP56/DDX39B is a major cotranscriptional RNA-DNA helicase that unwinds harmful R loops genome-wide. Genes Dev. 2020, 34(13-14): 898-912. DOI: 10.1101/gad.336024.119
Daley JM, Tomimatsu N, Hooks G, Wang W, Miller AS, Xue X, Nguyen KA, Kaur H, Williamson E, Mukherjee B, Hromas R, Burma S, Sung P. Specificity of end resection pathways for double-strand break regions containing ribonucleotides and base lesions. Nat Commun. 2020, 11(1): 3088. DOI: 10.1038/s41467-020-16903-4
Xue C, Daley JM, Xue X, Steinfeld J, Kwon Y, Sung P, Greene EC. Single-molecule visualization of human BLM helicase as it acts upon double- and single-stranded DNA substrates. Nucleic Acids Res. 2019, 47(21): 11225-11237. DOI: 10.1093/nar/gkz810
Jiang H, Xue X, Panda S, Kawale A, Hooy RM, Liang F, Sohn J, Sung P, Gekara NO. Chromatin-bound cGAS is an inhibitor of DNA repair and hence accelerates genome destabilization and cell death. EMBO J. 2019, 38(21): e102718. DOI: 10.15252/embj.2019102718
Wang W, Daley JM, Kwon Y, Xue X, Krasner D, Miller AS, Nguyen K, Williamson E, Shim E, Lee S, Hromas R, and Sung P. A DNA nick at Ku-blocked double-strand break ends serves as an entry site for exonuclease 1 (Exo1) or Sgs1-Dna2 in long-range DNA end resection. J Biol. Chem. 2018, 293(44):17061-17069. DOI: 10.1074/jbc.RA118.004769
Daley JM, Jimenez-Sainz J, Wang W, Miller AS, Xue X, Nguyen KA, Jensen RB, Sung P. Enhancement of BLM-DNA2-Mediated Long-Range DNA End Resection by CtIP. Cell Report. 2017, 21(2): 324-332. DOI: 10.1016/j.celrep.2017.09.048
Xue X, Papusha A, Choi K, Bonner J, Niu H, Kaur H, Zheng XF, Donnianni R, Lu L, Lichten M, Zhao X, Ira G, Sung P. Differential regulation of the anti-crossover and replication fork regression activities of Mph1 by Mte1. Genes Dev. 2016, 30(6): 687-699. DOI: 10.1101/gad.276139.115
Bonner J, Choi K, Xue X, Torres N, Szaka B, Wei L, Wan B, Arter M, Matos J, Sung P, Brown G, Branzei D, Zhao X. Smc5/6 Mediated Sumoylation of the Sgs1-Top3-Rmi1 Complex Promotes Removal of Recombination Intermediates. Cell Report. 2016, 16(2): 368-378. DOI: 10.1016/j.celrep.2016.06.015
Xue X, Sung P and Zhao X. Functions and regulation of multi-tasking FANCM family of DNA motor proteins (invited review). Genes Dev. 2015, 29(17): 1777-1788. DOI: 10.1101/gad.266593.115
Xue X*, Choi K* (co-first author), Bonner J, Szakal B, Papusha A, Saro D, Niu H, Ira G, Branzei D, Sung P, Zhao X. Selective modulation of the functions of a conserved DNA motor by a histone-fold complex. Genes Dev. 2015, 29(10): 1000-1005. DOI: 10.1101/gad.259143.115
Xue X, Choi K, Bonner J, Chiba T, Kwon Y, Xu Y, Sanchez H, Wyman C, Niu H, Zhao X, Sung P. Restriction of Replication Fork Regression Activities by a Conserved SMC Complex. Mol. Cell. 2014, 56(3): 436-445. DOI: 10.1016/j.molcel.2014.09.013
Zhao Q*, Xue X*, Longerich S* (co-first authors), Sung P, Xiong Y. Structural Insights into 5' Flap DNA Unwinding and Incision by the Human FAN1 Dimer. Nat Commun. 2014, 5:5726. DOI: 10.1038/ncomms6726
